Architecture 📐
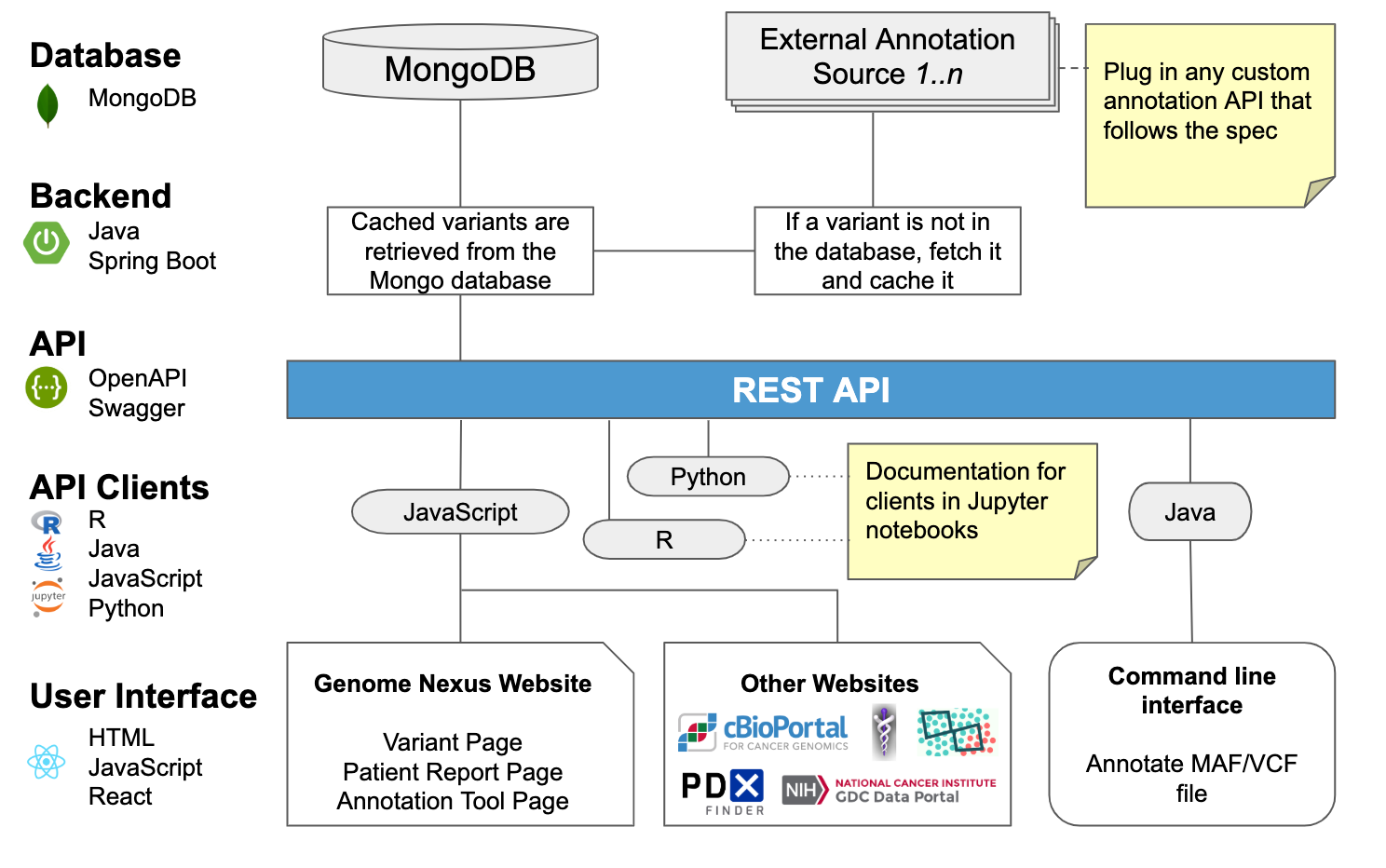
Backend
Genome Nexus aggregates variant annotation from various sources. There are two types:
Small sized annotations are stored as static data in the mongo database directly, see the genome-nexus-importer repo if you want to update/change this data.
Larger annotation sources are pulled on the fly from other APIs and cached in the mongo database.
For a list of all supported variant annotation sources see this Google Sheet:
https://docs.google.com/spreadsheets/d/1xML949NWzJGcvltjlquwSRIv79o13C_sfrVPAU5ci9Q/edit#gid=258442188
The code for the backend can be found here: https://github.com/genome-nexus/genome-nexus.
REST API
Genome Nexus provides a REST API for variant annotation. See API
API Clients
Clients in various languages can be generated to access it. See API
Genome Nexus Website
The frontend of the website is here: https://github.com/genome-nexus/genome-nexus-frontend.
Other Websites
The main consumer of the Genome Nexus REST API is cBioPortal. cBioPortal provides visualization, analysis, and download of large-scale cancer genomics data sets. Variants in cBioPortal are annotated using Genome Nexus. The other examples in the figure are potential other consumers. See also: Tools
Command Line Interface
There is a command line tool to annotate MAF and VCF files, see API
Last updated